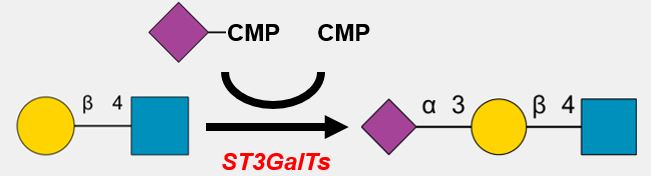
GlycoEnzDB
The Comprehensive Glyco-enzyme Database

| PAPS [Cyto] | → | PAPS [Golgi] |
* SLC35B2 and B3 transfer PAPS from the cytosol into the Golgi apparatus from the cytoplasm.
* PAPS is a universal sulfuryl donor for sulfation
* Depending on cancer type, the expression of PAPST1 and PAPST2 can increase or decrease, resulting in increases or decreases in sulfation of GAGs, respectively.
* Decreased expression of PAPST1 and PAPST2 caused FGF signaling to decrease in colorectal cells.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|

| miRNA Name | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|
| hsa-miR-642b-5p | 33-40 | -0.658 | 99.0 | -0.658 | 99.0 | noncon | |
| hsa-miR-6863 | 201-208 | -0.576 | 99.0 | -0.576 | 99.0 | noncon | |
| hsa-miR-21-3p | 376-383 | -0.554 | 99.0 | -0.53 | 99.0 | noncon | |
| hsa-miR-3591-3p | 376-383 | -0.519 | 99.0 | -0.497 | 99.0 | noncon | |
| hsa-miR-205-5p | 234-241 | -0.513 | 99.0 | -0.513 | 99.0 | con | |
| hsa-miR-4445-5p | 37-44 | -0.452 | 99.0 | -0.452 | 99.0 | noncon | |
| hsa-miR-6821-5p | 651-658 | -0.43 | 98.0 | -0.089 | 72.0 | noncon | |
| hsa-miR-2355-3p | 1184-1191 | -0.416 | 97.0 | -0.086 | 74.0 | noncon | |
| hsa-miR-3200-3p | 974-981 | -0.411 | 98.0 | -0.085 | 85.0 | noncon | |
| hsa-miR-371b-3p | 1626-1632 | -0.398 | 98.0 | -0.083 | 77.0 | noncon | |
| hsa-miR-548av-3p | 254-261 | -0.372 | 99.0 | -0.372 | 99.0 | noncon | |
| hsa-miR-431-5p | 294-300 | -0.365 | 98.0 | -0.365 | 98.0 | con | |
| hsa-miR-328-3p | 1426-1433 | -0.352 | 97.0 | -0.074 | 75.0 | noncon | |
| hsa-miR-6871-3p | 1446-1453 | -0.35 | 97.0 | -0.074 | 78.0 | noncon | |
| hsa-miR-9-5p | 79-85 | -0.343 | 98.0 | -0.343 | 99.0 | con | |
| hsa-miR-374a-3p | 70-77 | -0.342 | 99.0 | -0.342 | 99.0 | noncon | |
| hsa-miR-4686 | 345-351 | -0.336 | 97.0 | -0.333 | 98.0 | noncon | |
| hsa-miR-3978 | 57-64 | -0.336 | 99.0 | -0.336 | 99.0 | noncon | |
| hsa-miR-2114-5p | 65-71 | -0.335 | 98.0 | -0.335 | 98.0 | noncon | |
| hsa-miR-3657 | 1561-1568 | -0.326 | 94.0 | -0.069 | 69.0 | noncon | |
| hsa-miR-5007-3p | 124-131 | -0.324 | 98.0 | -0.324 | 99.0 | noncon | |
| hsa-miR-6836-3p | 91-97 | -0.323 | 97.0 | -0.323 | 98.0 | noncon | |
| hsa-miR-124-3p.2 | 2013-2020 | -0.322 | 92.0 | -0.068 | 60.0 | con | |
| hsa-miR-1279 | 230-237 | -0.322 | 99.0 | -0.322 | 99.0 | noncon | |
| hsa-miR-1255b-2-3p | 1945-1952 | -0.317 | 97.0 | -0.067 | 81.0 | noncon | |
| hsa-miR-6822-3p | 196-202 | -0.316 | 97.0 | -0.316 | 98.0 | noncon | |
| hsa-miR-4515 | 868-875 | -0.311 | 94.0 | -0.066 | 66.0 | noncon | |
| hsa-miR-3689c | 1492-1499 | -0.303 | 95.0 | -0.065 | 73.0 | noncon | |
| hsa-miR-3689a-3p | 1492-1499 | -0.303 | 95.0 | -0.065 | 73.0 | noncon | |
| hsa-miR-3689b-3p | 1492-1499 | -0.303 | 95.0 | -0.065 | 73.0 | noncon | |
| hsa-miR-3161 | 2100-2107 | -0.301 | 98.0 | -0.019 | 66.0 | noncon | |
| hsa-miR-1915-5p | 973-979 | -0.299 | 93.0 | -0.064 | 69.0 | noncon | |
| hsa-miR-6873-3p | 508-515 | -0.298 | 96.0 | -0.064 | 74.0 | noncon | |
| hsa-miR-6805-3p | 159-165 | -0.291 | 97.0 | -0.291 | 98.0 | noncon | |
| hsa-miR-5691 | 159-165 | -0.291 | 96.0 | -0.291 | 97.0 | noncon | |
| hsa-miR-1273h-5p | 1492-1499 | -0.287 | 93.0 | -0.062 | 70.0 | noncon | |
| hsa-miR-6779-5p | 1492-1499 | -0.287 | 94.0 | -0.062 | 71.0 | noncon | |
| hsa-miR-635 | 1045-1051 | -0.286 | 95.0 | -0.061 | 78.0 | noncon | |
| hsa-miR-4662a-5p | 304-310 | -0.285 | 96.0 | -0.285 | 98.0 | noncon | |
| hsa-miR-4755-5p | 32-38 | -0.284 | 97.0 | -0.284 | 98.0 | noncon | |
| hsa-miR-5006-3p | 32-38 | -0.284 | 97.0 | -0.284 | 98.0 | noncon | |
| hsa-miR-4275 | 1098-1105 | -0.284 | 98.0 | -0.061 | 88.0 | noncon | |
| hsa-miR-3616-3p | 1473-1480 | -0.282 | 92.0 | -0.06 | 56.0 | noncon | |
| hsa-miR-6511a-3p | 1365-1372 | -0.281 | 94.0 | -0.06 | 71.0 | noncon | |
| hsa-miR-6511b-3p | 1365-1372 | -0.281 | 94.0 | -0.06 | 71.0 | noncon | |
| hsa-miR-508-3p | 38-44 | -0.281 | 96.0 | -0.281 | 97.0 | noncon | |
| hsa-miR-508-3p | 1517-1524 | -0.275 | 95.0 | -0.059 | 65.0 | noncon | |
| hsa-miR-145-3p | 1061-1068 | -0.274 | 96.0 | -0.059 | 77.0 | noncon | |
| hsa-miR-199a-5p | 272-278 | -0.273 | 93.0 | -0.273 | 95.0 | con | |
| hsa-miR-199b-5p | 272-278 | -0.273 | 93.0 | -0.273 | 95.0 | con | |
| hsa-miR-1910-5p | 238-244 | -0.271 | 94.0 | -0.271 | 96.0 | noncon | |
| hsa-miR-30b-3p | 1492-1499 | -0.271 | 93.0 | -0.058 | 71.0 | noncon | |
| hsa-miR-6726-5p | 593-600 | -0.268 | 91.0 | -0.058 | 61.0 | noncon | |
| hsa-miR-4798-3p | 1760-1766 | -0.268 | 97.0 | -0.058 | 73.0 | noncon | |
| hsa-miR-6851-3p | 1425-1432 | -0.266 | 94.0 | -0.057 | 70.0 | noncon | |
| hsa-miR-5591-5p | 1437-1443 | -0.263 | 91.0 | -0.057 | 61.0 | noncon | |
| hsa-miR-4300 | 1437-1443 | -0.263 | 92.0 | -0.057 | 63.0 | noncon | |
| hsa-miR-206 | 363-369 | -0.262 | 94.0 | -0.252 | 95.0 | con | |
| hsa-miR-1-3p | 363-369 | -0.262 | 94.0 | -0.252 | 95.0 | con | |
| hsa-miR-6833-3p | 432-438 | -0.256 | 95.0 | -0.221 | 95.0 | noncon | |
| hsa-miR-613 | 363-369 | -0.251 | 93.0 | -0.241 | 95.0 | con | |
| hsa-miR-7702 | 1607-1614 | -0.25 | 93.0 | -0.054 | 69.0 | noncon | |
| hsa-miR-6780a-5p | 1492-1499 | -0.25 | 92.0 | -0.054 | 69.0 | noncon |