GlycoEnzDB
The Comprehensive Glyco-enzyme Database

.jpg)
|
+ |
GlcNAc(b1-0)R(-CHAR).jpg)
|
→ |
Gal(b1-4)GlcNAc(b1-0)R(-CHAR).jpg)
|
+ | UDP |
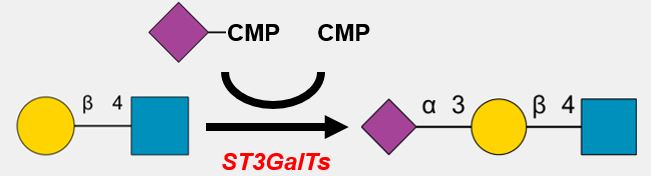
* Synthesizes polylacosamine of N-glycan, particulalry along the GlcNAc(β1-6) N-glycan branch.
* B3GnT2 and B3GnT8 may form a heterocomplex to facilitate polyLacNAc extension.
* B3GNT8 is highly expressed in lung, throat, and ileum. It is further augmented during colon cancer.
* Highly metastatic cell lines may synthesize more N-glycans containing polylactosamines than poorly metastatic cell lines.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|

| miRNA Name | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|
| hsa-miR-759 | 219-226 | -0.516 | 99.0 | -0.378 | 99.0 | noncon | |
| hsa-miR-7111-5p | 134-141 | -0.405 | 94.0 | -0.405 | 95.0 | noncon | |
| hsa-miR-4745-5p | 217-223 | -0.401 | 97.0 | -0.297 | 95.0 | noncon | |
| hsa-miR-6806-5p | 184-190 | -0.398 | 94.0 | -0.398 | 96.0 | noncon | |
| hsa-miR-4723-5p | 134-141 | -0.397 | 94.0 | -0.397 | 95.0 | noncon | |
| hsa-miR-4481 | 217-223 | -0.394 | 98.0 | -0.292 | 96.0 | noncon | |
| hsa-miR-6763-5p | 214-220 | -0.391 | 98.0 | -0.391 | 98.0 | noncon | |
| hsa-miR-5698 | 134-141 | -0.389 | 94.0 | -0.389 | 95.0 | noncon | |
| hsa-miR-3065-3p | 21-28 | -0.382 | 98.0 | -0.382 | 99.0 | noncon | |
| hsa-miR-6870-5p | 134-141 | -0.381 | 93.0 | -0.381 | 94.0 | noncon | |
| hsa-miR-491-5p | 215-221 | -0.377 | 97.0 | -0.28 | 95.0 | noncon | |
| hsa-miR-3150a-3p | 214-220 | -0.369 | 97.0 | -0.369 | 98.0 | noncon | |
| hsa-miR-504-3p | 211-217 | -0.352 | 98.0 | -0.352 | 98.0 | noncon | |
| hsa-miR-1296-3p | 216-222 | -0.349 | 98.0 | -0.26 | 97.0 | noncon | |
| hsa-miR-3175 | 214-220 | -0.343 | 96.0 | -0.343 | 97.0 | noncon | |
| hsa-miR-3661 | 74-80 | -0.342 | 96.0 | -0.342 | 97.0 | noncon | |
| hsa-miR-4666b | 207-213 | -0.34 | 97.0 | -0.34 | 98.0 | noncon | |
| hsa-miR-4505 | 138-145 | -0.334 | 96.0 | -0.334 | 97.0 | noncon | |
| hsa-miR-631 | 74-80 | -0.321 | 95.0 | -0.321 | 96.0 | noncon | |
| hsa-miR-132-3p | 175-181 | -0.314 | 98.0 | -0.314 | 98.0 | noncon | |
| hsa-miR-5008-5p | 37-43 | -0.285 | 94.0 | -0.285 | 96.0 | noncon | |
| hsa-miR-212-3p | 175-181 | -0.281 | 97.0 | -0.281 | 98.0 | noncon | |
| hsa-miR-5787 | 138-145 | -0.264 | 96.0 | -0.264 | 97.0 | noncon | |
| hsa-miR-455-3p.1 | 173-179 | -0.261 | 96.0 | -0.261 | 97.0 | noncon | |
| hsa-miR-1199-5p | 58-64 | -0.259 | 95.0 | -0.259 | 96.0 | noncon |