SLC35D1
General Information
Protein name:
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter
Reaction (SNFG):
|
UDP-GlcA [cyto] or UDP-GalNAc [cyto] or UDP-GlcA [cyto] or UDP-GalNAc [cyto]
|
→
|
UDP-GlcA [ER] or UDP-GalNAc [ER] or UDP-GlcA [Golgi] or UDP-GalNAc [Golgi]
|
Reaction (IUBMB):
No specified reaction.
Related Pathway
Related Database
Enzyme Specificity
Substrate:
UDP-GlcA [cyto] or UDP-GalNAc [cyto] or UDP-GlcA [cyto] or UDP-GalNAc [cyto](-CHAR)
Product:
UDP-GlcA [ER] or UDP-GalNAc [ER] or UDP-GlcA [Golgi] or UDP-GalNAc [Golgi](-CHAR)
Simulation Parameters
Mechaelis-Menten
parameters (suggested)
Vm: (pmol/cell • hr)
Km:
(pM)
Relative conc of
enzymes in compartment.(sum will be normalized to 1)
Expression data are collected from NCBI gene database
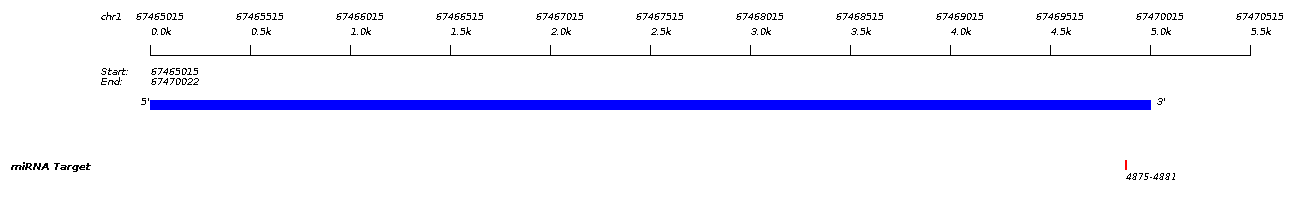
| miRNA Target
| Site Type
| Pos.
| Context Score
| Frac.
| Wt. Context
| Frac.
| Conserved
| Quality
|
5'...GGUACUGAGGAAUU
GAGGUAU
G...
3' GUGGACUAUUUUGA
CUCCAUA
U
|
7mer-m8 |
4875-4881 |
-0.253 |
94.0 |
-0.038 |
72.0 |
noncon |
|