GlycoEnzDB
The Comprehensive Glyco-enzyme Database

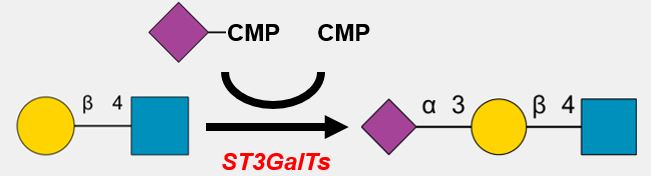
| PAPS [Cyto] | → | PAPS [Golgi] |
* SLC35B2 and B3 transfer PAPS from the cytosol into the Golgi apparatus from the cytoplasm.
* PAPS is a universal sulfuryl donor for sulfation.
* Depending on cancer type, the expression of PAPST1 and PAPST2 can increase or decrease, resulting in increases or decreases in sulfation of GAGs, respectively.
* Decreased expression of PAPST1 and PAPST2 caused FGF signaling to decrease in colorectal cells.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|
| SLC35B2/hsa-miR-2467-3p |
|---|
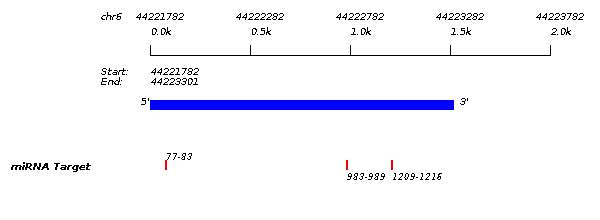
| miRNA Target | Site Type | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|---|
|
5'...UCCAGACUCACUCC
CCUCUGCA
... 3' GGACUCGGAGAGAC GGAGACG A |
8mer | 1209-1216 | -0.456 | 99.0 | -0.454 | 99.0 | noncon | |
|
5'...GCCUCUCCUGUGUU
CUCUGCA
A... 3' GGACUCGGAGAGACG GAGACG A |
7mer-A1 | 77-83 | -0.263 | 95.0 | -0.263 | 96.0 | noncon | |
|
5'...GUAUCACAGACCAG
CUCUGCA
G... 3' GGACUCGGAGAGACG GAGACG A |
7mer-A1 | 983-989 | -0.089 | 68.0 | -0.089 | 78.0 | noncon |