GlycoEnzDB
The Comprehensive Glyco-enzyme Database

| GlcN(a1-6)(ancyl)PI | → | Man(a1-4)GlcN(a1-6)(ancyl)PI |
* PIGM together with PIGX mediated transfer of the first Man from Dol-P (donor) during GPI-protein synthesis.
* Here, PIGM has catalytic activity while PIGX has regulatory function.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|
| PIGM/hsa-miR-4433b-5p |
|---|
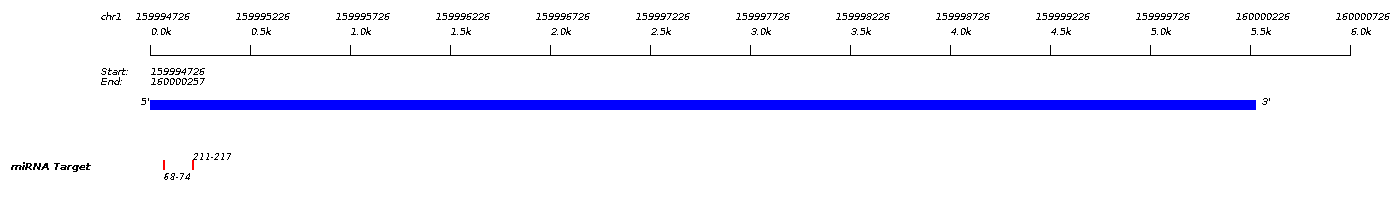
| miRNA Target | Site Type | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|---|
|
5'...CAGAAGAGAGCUU
UGGGACA
U... 3' UGUCCUCACCCCC ACCCUGU A |
7mer-m8 | 68-74 | -0.256 | 92.0 | -0.256 | 94.0 | noncon | |
|
5'...GGACUCGUUUAUU
UGGGACA
U... 3' UGUCCUCACCCCC ACCCUGU A |
7mer-m8 | 211-217 | -0.216 | 88.0 | -0.205 | 90.0 | noncon |