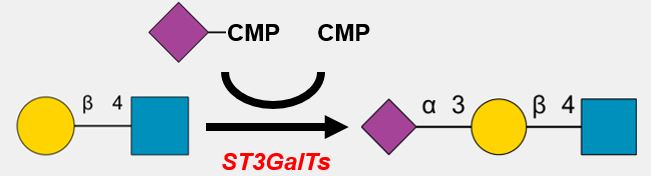
GlycoEnzDB
The Comprehensive Glyco-enzyme Database

GlcNAc(b1-0)AsnR(-CHAR).jpg)
|
→ |
AsnR(-CHAR).jpg)
|
* Involved in the lysosomal degradation of N-linked glycans in the lysosome.
* Cleaves GlcNAc(b1-4)GlcNAc(b1-?)Asn.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|

| miRNA Name | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|
| hsa-miR-2114-3p | 697-704 | -0.527 | 99.0 | -0.423 | 98.0 | noncon | |
| hsa-miR-6835-5p | 1427-1434 | -0.511 | 99.0 | -0.411 | 98.0 | noncon | |
| hsa-miR-6823-3p | 697-704 | -0.49 | 99.0 | -0.395 | 98.0 | noncon | |
| hsa-miR-6515-5p | 432-439 | -0.453 | 99.0 | -0.366 | 98.0 | noncon | |
| hsa-let-7e-3p | 97-103 | -0.442 | 95.0 | -0.442 | 97.0 | noncon | |
| hsa-miR-625-5p | 423-430 | -0.424 | 97.0 | -0.343 | 95.0 | noncon | |
| hsa-miR-7515 | 292-299 | -0.423 | 99.0 | -0.342 | 98.0 | noncon | |
| hsa-miR-212-5p | 1793-1800 | -0.423 | 98.0 | -0.342 | 98.0 | con | |
| hsa-miR-4665-5p | 424-431 | -0.415 | 97.0 | -0.336 | 96.0 | noncon | |
| hsa-miR-4790-3p | 1686-1693 | -0.4 | 99.0 | -0.324 | 98.0 | noncon | |
| hsa-miR-3115 | 4098-4105 | -0.374 | 96.0 | -0.051 | 59.0 | noncon | |
| hsa-miR-196b-5p | 663-670 | -0.37 | 97.0 | -0.3 | 97.0 | noncon | |
| hsa-miR-196a-5p | 663-670 | -0.37 | 97.0 | -0.3 | 96.0 | noncon | |
| hsa-miR-4703-3p | 679-686 | -0.362 | 97.0 | -0.294 | 97.0 | noncon | |
| hsa-miR-3649 | 3962-3969 | -0.362 | 96.0 | -0.049 | 56.0 | noncon | |
| hsa-miR-643 | 4948-4955 | -0.361 | 98.0 | -0.049 | 70.0 | noncon | |
| hsa-miR-3691-3p | 1666-1673 | -0.357 | 98.0 | -0.29 | 97.0 | noncon | |
| hsa-miR-1267 | 44-51 | -0.352 | 98.0 | -0.352 | 98.0 | noncon | |
| hsa-miR-433-3p | 4671-4678 | -0.338 | 98.0 | -0.046 | 76.0 | con | |
| hsa-miR-6751-5p | 1428-1434 | -0.336 | 96.0 | -0.273 | 95.0 | noncon | |
| hsa-miR-223-5p | 150-157 | -0.333 | 96.0 | -0.271 | 96.0 | noncon | |
| hsa-miR-6853-3p | 1472-1479 | -0.328 | 97.0 | -0.267 | 97.0 | noncon | |
| hsa-miR-3916 | 219-226 | -0.326 | 98.0 | -0.266 | 98.0 | noncon | |
| hsa-miR-1275 | 424-431 | -0.318 | 96.0 | -0.259 | 96.0 | noncon | |
| hsa-miR-5004-5p | 162-169 | -0.317 | 95.0 | -0.258 | 94.0 | noncon | |
| hsa-miR-6733-3p | 1016-1023 | -0.308 | 97.0 | -0.251 | 96.0 | noncon | |
| hsa-miR-3125 | 219-226 | -0.308 | 98.0 | -0.251 | 97.0 | noncon | |
| hsa-miR-6773-3p | 1297-1304 | -0.304 | 96.0 | -0.248 | 95.0 | noncon | |
| hsa-miR-6864-5p | 293-299 | -0.302 | 96.0 | -0.246 | 95.0 | noncon | |
| hsa-miR-4303 | 1408-1415 | -0.3 | 97.0 | -0.245 | 97.0 | noncon | |
| hsa-miR-3616-3p | 5314-5320 | -0.3 | 93.0 | -0.041 | 46.0 | noncon | |
| hsa-miR-503-3p | 1739-1746 | -0.295 | 95.0 | -0.241 | 95.0 | noncon | |
| hsa-miR-516a-5p | 3353-3360 | -0.295 | 92.0 | -0.041 | 54.0 | noncon | |
| hsa-miR-4766-5p | 266-273 | -0.293 | 99.0 | -0.239 | 99.0 | noncon | |
| hsa-miR-875-3p | 1445-1452 | -0.292 | 98.0 | -0.238 | 97.0 | noncon | |
| hsa-miR-630 | 4059-4066 | -0.289 | 95.0 | -0.04 | 59.0 | noncon | |
| hsa-miR-6803-5p | 1428-1434 | -0.289 | 94.0 | -0.236 | 93.0 | noncon | |
| hsa-miR-518a-5p | 1545-1552 | -0.288 | 98.0 | -0.235 | 98.0 | noncon | |
| hsa-miR-527 | 1545-1552 | -0.288 | 98.0 | -0.235 | 98.0 | noncon | |
| hsa-miR-4320 | 591-598 | -0.285 | 96.0 | -0.233 | 95.0 | noncon | |
| hsa-miR-448 | 4929-4936 | -0.285 | 97.0 | -0.04 | 69.0 | noncon | |
| hsa-miR-4782-5p | 594-601 | -0.285 | 97.0 | -0.233 | 96.0 | noncon | |
| hsa-miR-4638-3p | 4990-4997 | -0.28 | 94.0 | -0.039 | 57.0 | noncon | |
| hsa-miR-4528 | 1469-1476 | -0.28 | 99.0 | -0.229 | 98.0 | noncon | |
| hsa-miR-668-3p | 3035-3042 | -0.279 | 95.0 | -0.039 | 72.0 | noncon | |
| hsa-miR-5706 | 594-601 | -0.277 | 96.0 | -0.226 | 96.0 | noncon | |
| hsa-miR-4660 | 987-994 | -0.276 | 94.0 | -0.226 | 93.0 | noncon | |
| hsa-miR-6507-3p | 5167-5174 | -0.274 | 97.0 | -0.038 | 79.0 | noncon | |
| hsa-miR-4309 | 521-527 | -0.274 | 95.0 | -0.224 | 95.0 | noncon | |
| hsa-miR-610 | 1569-1575 | -0.271 | 94.0 | -0.221 | 93.0 | noncon | |
| hsa-miR-4308 | 1107-1114 | -0.269 | 97.0 | -0.22 | 96.0 | noncon | |
| hsa-miR-542-3p | 759-766 | -0.268 | 96.0 | -0.219 | 95.0 | noncon | |
| hsa-miR-4507 | 5292-5299 | -0.266 | 94.0 | -0.037 | 66.0 | noncon | |
| hsa-miR-95-5p | 4384-4390 | -0.263 | 98.0 | -0.037 | 73.0 | noncon | |
| hsa-miR-154-5p | 4680-4687 | -0.261 | 96.0 | -0.036 | 66.0 | con | |
| hsa-miR-6514-5p | 522-529 | -0.26 | 96.0 | -0.213 | 96.0 | noncon | |
| hsa-miR-3118 | 3478-3485 | -0.259 | 93.0 | -0.036 | 58.0 | noncon | |
| hsa-miR-3064-3p | 2395-2402 | -0.259 | 92.0 | -0.036 | 59.0 | noncon | |
| hsa-miR-1244 | 3425-3432 | -0.25 | 93.0 | -0.035 | 59.0 | noncon |