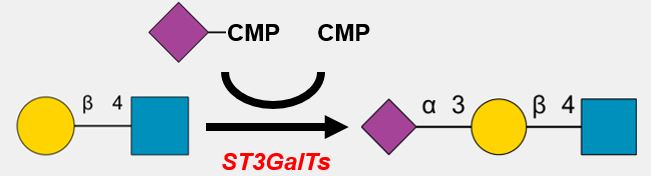

.jpg)
|
+ |
R(-CHAR).jpg)
|
→ |
Gal(b1-0)R(-CHAR).jpg)
|
+ | UDP |
* Transfers GlcNAc(α1-4) to βGal preferentially present in O-glycans forming GlcNAc(α1-4)Galβ-R or terminal αGlcNAc.
* The enzyme shows preference to act on core-2 glycan rather than core-1 glycans indicating a role for GlcNAc(β1-6) in promoting its activity. It extends the core-1 arm of O-glycans.
* It can act on both arms of core-2 glycan to corm structures like GlcNAc(α1-4)Gal(β1-4)GlcNAc(β1-6)[GlcNAc(α1-4)Gal(β1-3)]GalNAc.
* A4GNT is expressed in stomach, gall bladder, duodenum, and thus such structures are normally restricted to gland & gastric mucins, particulalry MUC6 with a small amount also being associated with MUC5AC.
* αGlcNAc is also expressed in pancreatic ducts showing gastric metaplasia or pancreatic intraepithelial neoplasia 1 (PanIN-1).
* A4gnt knockout mice revealed that αGlcNAc serves as a tumor suppressor of differentiated type of gastric adenocarcinoma. In addition, αGlcNAc protects gastric gland mucous cells from H. pylori infection. Thus, αGlcNAc plays a dual role to prevent the gastric mucosa from gastric tumorigenesis.
| ER | Cis- | Medial- | Trans- | TGN |
|---|---|---|---|---|

| miRNA Name | Pos. | Context Score | Frac. | Wt. Context | Frac. | Conserved | Quality |
|---|---|---|---|---|---|---|---|
| hsa-let-7e-3p | 302-309 | -0.723 | 99.0 | -0.723 | 99.0 | noncon | |
| hsa-miR-3136-5p | 524-531 | -0.607 | 99.0 | -0.607 | 99.0 | noncon | |
| hsa-miR-22-3p | 397-404 | -0.601 | 99.0 | -0.601 | 99.0 | con | |
| hsa-miR-6772-5p | 182-189 | -0.569 | 99.0 | -0.569 | 99.0 | noncon | |
| hsa-miR-6769a-5p | 216-223 | -0.56 | 99.0 | -0.56 | 99.0 | noncon | |
| hsa-miR-4777-3p | 470-477 | -0.549 | 99.0 | -0.549 | 99.0 | noncon | |
| hsa-miR-575 | 167-174 | -0.54 | 99.0 | -0.54 | 99.0 | noncon | |
| hsa-miR-891a-3p | 52-59 | -0.534 | 99.0 | -0.534 | 99.0 | noncon | |
| hsa-miR-6769b-5p | 216-223 | -0.534 | 99.0 | -0.534 | 99.0 | noncon | |
| hsa-miR-660-5p | 381-388 | -0.53 | 99.0 | -0.53 | 99.0 | noncon | |
| hsa-miR-4439 | 524-531 | -0.526 | 99.0 | -0.526 | 99.0 | noncon | |
| hsa-miR-4676-5p | 167-174 | -0.516 | 99.0 | -0.516 | 99.0 | noncon | |
| hsa-miR-148b-3p | 325-332 | -0.498 | 99.0 | -0.498 | 99.0 | noncon | |
| hsa-miR-152-3p | 325-332 | -0.498 | 99.0 | -0.498 | 99.0 | noncon | |
| hsa-miR-148a-3p | 325-332 | -0.498 | 99.0 | -0.498 | 99.0 | noncon | |
| hsa-miR-4683 | 69-76 | -0.464 | 99.0 | -0.464 | 99.0 | noncon | |
| hsa-miR-6513-5p | 155-162 | -0.461 | 99.0 | -0.461 | 99.0 | noncon | |
| hsa-miR-3619-3p | 189-196 | -0.451 | 99.0 | -0.451 | 99.0 | noncon | |
| hsa-miR-6881-3p | 254-261 | -0.439 | 98.0 | -0.439 | 99.0 | noncon | |
| hsa-miR-1301-3p | 342-349 | -0.415 | 98.0 | -0.415 | 99.0 | noncon | |
| hsa-miR-5047 | 342-349 | -0.415 | 98.0 | -0.415 | 98.0 | noncon | |
| hsa-miR-567 | 178-185 | -0.412 | 99.0 | -0.412 | 99.0 | noncon | |
| hsa-miR-4264 | 331-338 | -0.412 | 99.0 | -0.412 | 99.0 | noncon | |
| hsa-miR-5089-5p | 154-161 | -0.401 | 99.0 | -0.401 | 99.0 | noncon | |
| hsa-miR-873-5p.2 | 290-297 | -0.381 | 99.0 | -0.381 | 99.0 | noncon | |
| hsa-miR-7977 | 355-362 | -0.361 | 99.0 | -0.361 | 99.0 | noncon | |
| hsa-miR-6875-3p | 372-379 | -0.358 | 98.0 | -0.358 | 99.0 | noncon | |
| hsa-miR-4455 | 183-189 | -0.332 | 91.0 | -0.332 | 94.0 | noncon | |
| hsa-miR-6882-3p | 396-402 | -0.322 | 96.0 | -0.322 | 97.0 | noncon | |
| hsa-miR-4274 | 200-206 | -0.318 | 97.0 | -0.318 | 98.0 | noncon | |
| hsa-miR-4712-5p | 386-392 | -0.306 | 96.0 | -0.306 | 97.0 | noncon | |
| hsa-miR-5591-3p | 382-388 | -0.302 | 96.0 | -0.302 | 97.0 | noncon | |
| hsa-miR-500a-3p | 262-268 | -0.3 | 96.0 | -0.3 | 97.0 | noncon | |
| hsa-miR-6503-3p | 493-499 | -0.296 | 96.0 | -0.296 | 97.0 | noncon | |
| hsa-miR-770-5p | 386-392 | -0.295 | 96.0 | -0.295 | 97.0 | noncon | |
| hsa-miR-4264 | 467-473 | -0.292 | 97.0 | -0.292 | 97.0 | noncon | |
| hsa-miR-6855-3p | 524-530 | -0.29 | 96.0 | -0.29 | 97.0 | noncon | |
| hsa-miR-1324 | 138-144 | -0.29 | 97.0 | -0.29 | 98.0 | noncon | |
| hsa-miR-4776-5p | 190-196 | -0.289 | 95.0 | -0.289 | 96.0 | noncon | |
| hsa-miR-3184-3p | 481-487 | -0.282 | 97.0 | -0.282 | 98.0 | noncon | |
| hsa-miR-6792-3p | 369-375 | -0.279 | 95.0 | -0.279 | 96.0 | noncon | |
| hsa-miR-4258 | 85-91 | -0.275 | 91.0 | -0.275 | 93.0 | noncon | |
| hsa-miR-6780a-3p | 255-261 | -0.273 | 96.0 | -0.273 | 97.0 | noncon | |
| hsa-miR-200b-5p | 435-441 | -0.269 | 97.0 | -0.269 | 98.0 | noncon | |
| hsa-miR-200a-5p | 435-441 | -0.269 | 97.0 | -0.269 | 98.0 | noncon | |
| hsa-miR-4697-3p | 327-333 | -0.267 | 92.0 | -0.267 | 94.0 | noncon | |
| hsa-miR-4513 | 524-530 | -0.266 | 95.0 | -0.266 | 96.0 | noncon | |
| hsa-miR-3121-3p | 422-429 | -0.265 | 99.0 | -0.265 | 99.0 | noncon | |
| hsa-miR-6804-3p | 262-268 | -0.263 | 92.0 | -0.263 | 95.0 | noncon | |
| hsa-miR-609 | 183-189 | -0.261 | 92.0 | -0.261 | 94.0 | noncon | |
| hsa-miR-627-3p | 250-257 | -0.26 | 97.0 | -0.26 | 98.0 | noncon | |
| hsa-miR-7153-3p | 187-193 | -0.258 | 96.0 | -0.258 | 97.0 | noncon | |
| hsa-miR-450a-1-3p | 156-162 | -0.257 | 96.0 | -0.257 | 97.0 | noncon |